About me

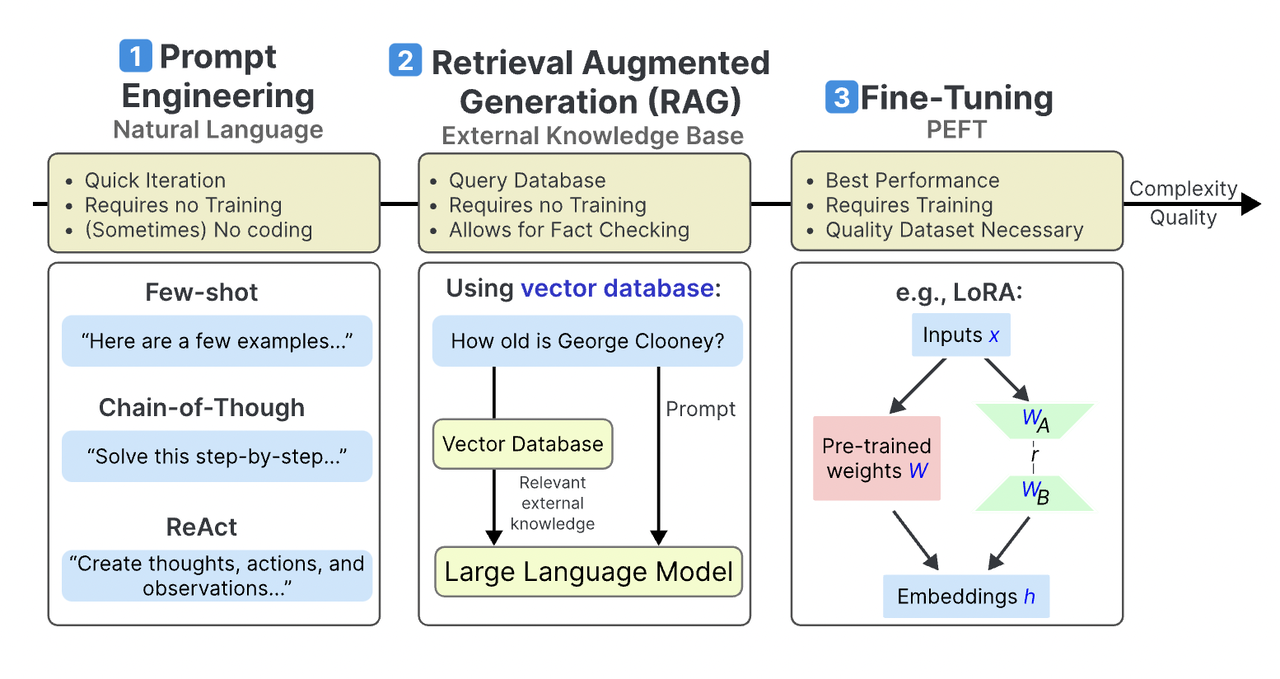
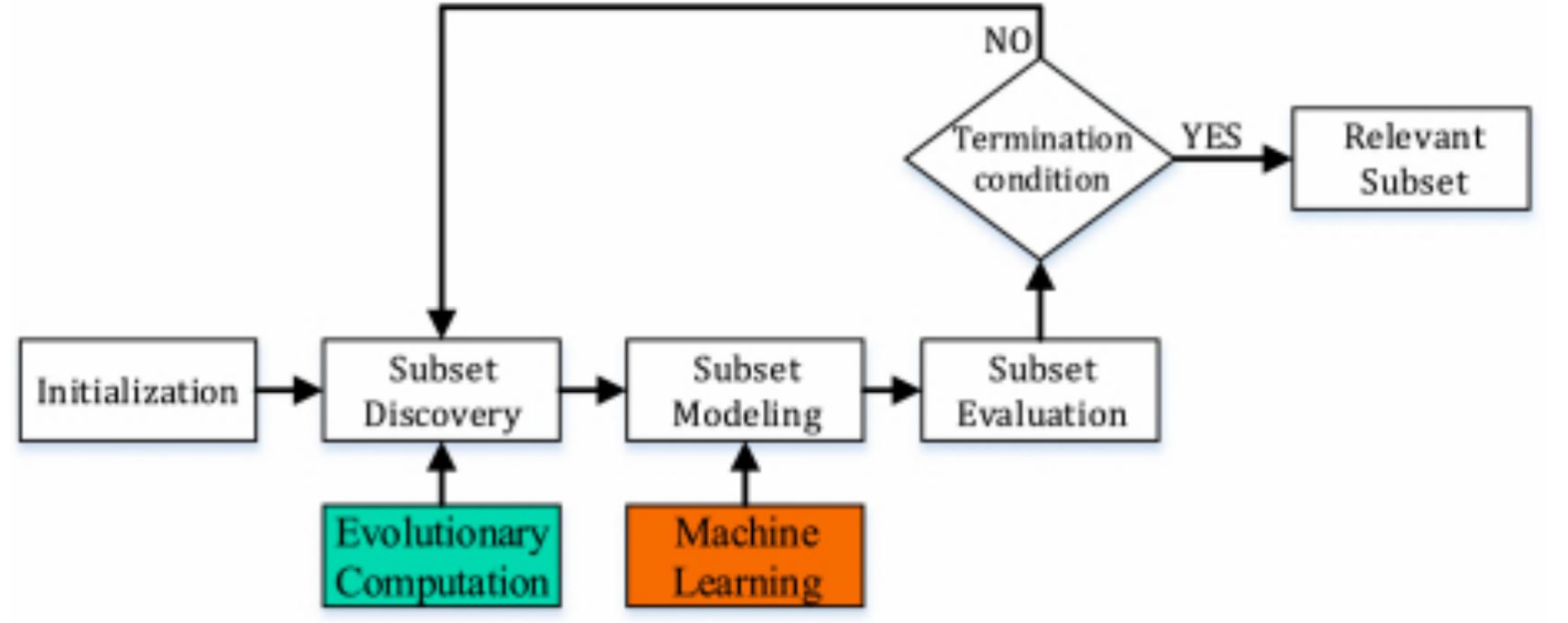
Welcome to my page! My research interests include advanced machine learning, AI techniques, data analysis, and dynamic optimization. I develop novel approaches in machine learning, focusing on foundation models and their scientific applications. My expertise encompasses data mining for high-dimensional data, feature selection, and knowledge extraction from large datasets, along with optimizing AI models for better efficiency and performance. I have published in top conferences and journals, such as ICML, TKDE, and CSUR, and my work has been featured by MIT Technology Review. Additionally, my knowledge base construction and management system has garnered more than 200 stars on GitHub, reflecting its value and utility within the community. By leveraging interdisciplinary knowledge in AI, data science, and algorithm design, I aim to achieve breakthroughs in machine intelligence that benefit various scientific and practical domains. You can reach me at email: wangxubin [at] kindlab [dot] site. Any information or suggestions would be greatly appreciated.